A transposon, also known as a transposable element or jumping gene, is a DNA sequence that can change its position within the genome. Sometimes, it creates or reverses mutations that alter the genetic characteristics of cells and the size of the genome. Transposons typically result in the duplication of the same genetic material.
Generally, transposons can be classified into three main groups based on their mode of transposition: Type I transposons (Class I elements), Type II transposons (Class II elements), and Helitron transposons.
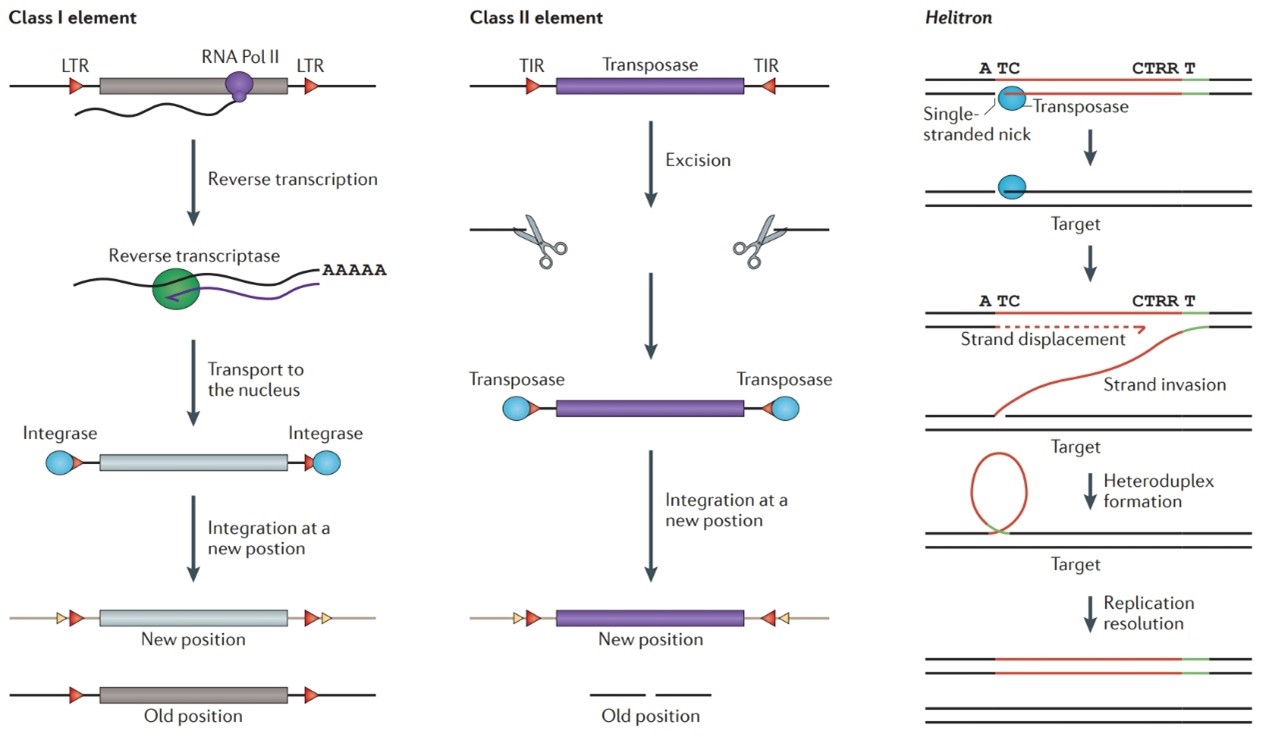
Fig 1. 3 types of transposons
Type I transposons are also called retrotransposons. According to their transposition mechanism, they are figuratively referred to as "copy-paste" type transposons. When a retrotransposon transposes, it first uses DNA as a template and transcribes it into mRNA under the action of RNA polymerase II. Then, this mRNA is used as a template for reverse transcription into cDNA, which is integrated into a new location on the genome under the action of integrase enzyme.
Type II transposons are simply called transposons. Unlike retrotransposons, the transposition mechanism of Type II transposons is known as "cut-and-paste." Under the action of the enzyme transposase, a Type II transposon is dissociated from its original position and reintegrated into the chromosome. The broken strand at the original position due to transposon dissociation is repaired by the DNA repair mechanism. As a result, the original position has one less transposon sequence, while the new position has one more. Like retrotransposons, Type II transposons can be categorized into autonomous and non-autonomous types. Non-autonomous transposons lack all the components necessary for transposition and are therefore dependent on autonomous transposons. Common Type II transposons include Tn3, Tn5, and Tn10 from bacteria; piggyBac (PB) and Mariner transposons from insects; and Sleeping Beauty (SB) and Tol2 transposons from fish.
Helitron transposons are a new type of DNA transposon discovered in recent years. They were initially identified in the Arabidopsis genome using computational methods based on repetitive sequences. Later, it was found that most plants and many animal genomes carry Helitron transposons. These transposons have typical 5'TC and 3'CTRR (where R is either A or G) termini and a stem-loop structure about 15 to 20 bp upstream of the 3' terminus, which serves as the termination signal of the transposon. After transposition, Helitrons are usually inserted into the AT target site in the AT-rich region. Unlike retrotransposons and traditional transposons, Helitrons transpose by rolling circles. Moreover, they often capture and carry gene fragments during the process of rolling circle replication, which can lead to changes in gene copy number and promote genome evolution to a certain extent.
Tn5 Transposon
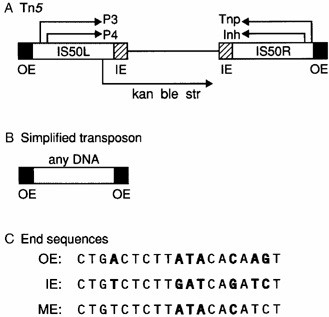
Fig2. Structure of Tn5 transposon
The Tn5 transposon consists of core sequences encoding three resistance genes (kan, ble, str) and two inverted IS50 sequences. The sequences of IS50R and IS50L are highly homologous and contain genes encoding a transposase (TnP) and a transposition inhibitory protein (lnh). However, there is a mutation in one of the bases in IS50L that causes premature termination of translation. Thus, only IS50R produces normal active TnP and lnh. IS50 has a 19bp inverted end (outer end OE and inner end IE), and the two inverted ends differ by 7 base pairs (bolded bases in Fig 2. This inverted end is the site of action of the transposase enzyme (Tnp). The IE is recognized by Tnp only in the absence of Dam-DNA methylation. ME is a chimera of IE and OE. Both OE and IE are suboptimal sequences, while ME is the hyperactive sequence.
OE: CTGACTCTTATACACAAGT
IE: CTGTCTCTTGATCAGATCT
ME: CTGTCTCTTATACACATCT
Researchers have found that the entire transposon sequence is not necessary for transposition. Only the end core sequence of the transposon (OE/IE/ME) is required. Then the transposase can insert and ligate this part of the sequence into the genome (Fig 3). The insertion site of Tn5 transposase is highly randomized, making it widely used in the fields of in vitro transgenesis (integration of exogenous genes into host cells) and second-generation sequencing for library construction. Tnp binding and insertion also show some preference. Their preferred DNA target sequence is A-GNT(T/C)(A/T)(A/G)ANC-T, where N is any nucleotide.
Wild-type Tnp is a very inactive protein. Purified wild-type Tnp has no transposition activity in vitro and a very low transposition frequency in vivo. Therefore, in practical applications, mutant forms of Tn5 transposase are generally used to enhance the activity of Tnp, such as L372P, E54K, E110K, E345K, P242A, or P242G.
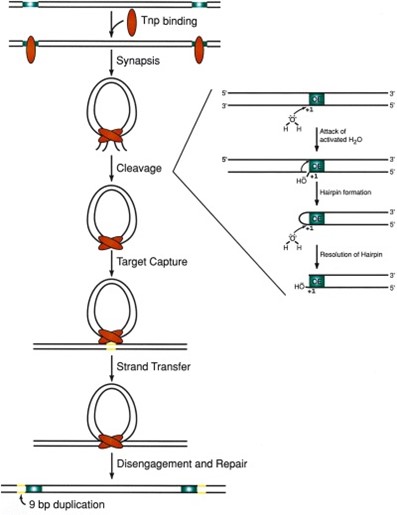
Fig 3. The principle of Tn5 transposition
PiggyBac Transposition System
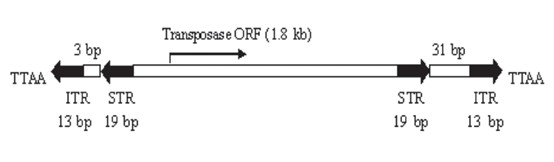
Fig 4. Structure of PB transposition system
The full length of PB is 2472 bp. At each end, there is an inverted terminal repeat (ITR) of 13 bp and 19 bp. In the middle, it encodes a transposase with 594 amino acid residues. The two ends of the transposon are differentiated according to the transcription direction of the transposase. At the outermost part of each end, there is a 13bp-long ITR that is symmetrical. Inside, there is a spacer region at each end that is asymmetrical (3 bp on the left end and 31 bp on the right end). Further inside, there is a 19bp-long and symmetrical sub-terminal inverted repeat (STR). At the 5' end of the inverted repeat sequence, there are 2 to 3 C bases. The corresponding G bases at the 3' end play a role in the selection of the shear site. It was found that only PB transposons with the left LTR longer than 311 bp and the right LTR longer than 235 bp have transposition activity. The combination study of the left and right ends of the transposon shows that only "left+right" and "right+left" have transposition activity. The transposition activity of the former is 4.6 times that of the latter.
After the discovery of the PiggyBac transposon, it underwent a series of optimization and modification processes, such as the optimization and mutation of the transposase codon and the simplification of the two-terminal ITR sequences. Finally, a complete PiggyBac vector system was formed. This system consists of a helper plasmid encoding the transposase enzyme and a transposon plasmid. The transposon plasmid contains optimized sub-terminal inverse repeat sequences at both ends and a transposable region in the middle. Researchers can insert the target gene sequence they want to transpose into the host genome into this region.
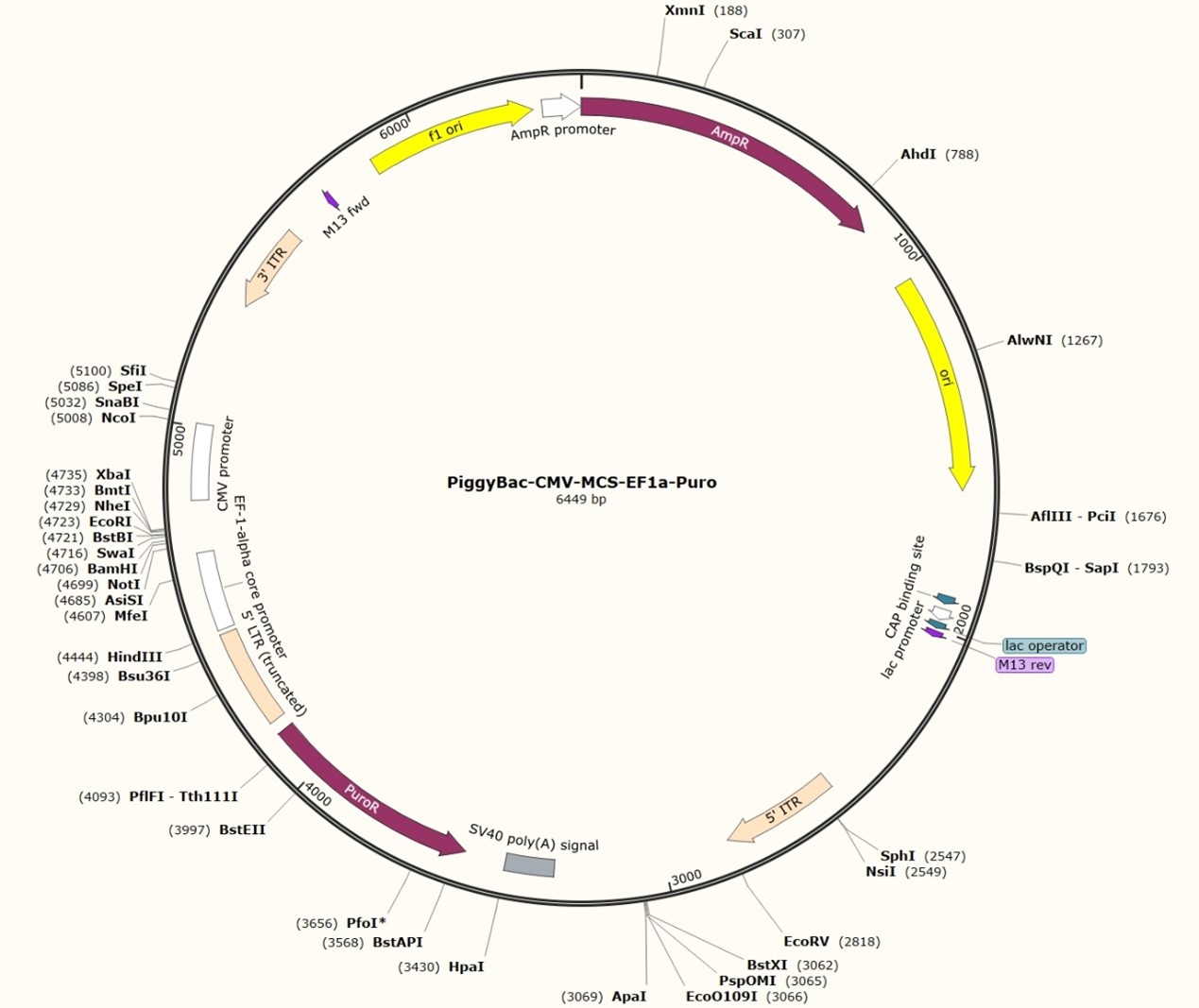
Fig 5. A typical PiggyBac transposon plasmid map
In the presence of PiggyBac transposase, the 5'ITR and 3'ITR and the DNA fragments between them are integrated into the genomic locus containing the TTAA sequence.
In the experiment, the helper plasmid and the transposon plasmid are co-transformed into the target cells. The transposase encoded by the helper plasmid recognizes the ITR sequences at both ends of the transposon plasmid and cleaves them, releasing the transposable region. The transposase then integrates this region into the TTAA-containing sites in the host genome. At the same time, TTAA repeat sequences appear at both ends of the transposed region. PiggyBac can transpose a fragment of up to 20 kilobases in size.
Sleeping Beauty Transposon and Tol2 Transposon
The "Sleeping Beauty" transposon is a member of the Tc1/mariner transposon superfamily. It was originally a natural genetic component of vertebrates such as humans, animals, and fish. However, over hundreds of millions of years of evolution, most of these transposons became inactive. In 1997, Ivics et al. from the University of Minnesota collected the sequences of 12 inactivated Tc1-like transposase genes from eight fish species. They performed multiple sequence comparisons and molecularly reconstructed these genes to restore their jumping ability. This study reactivated transposons that had been dormant for more than 10 million years, hence the name "Sleeping Beauty."
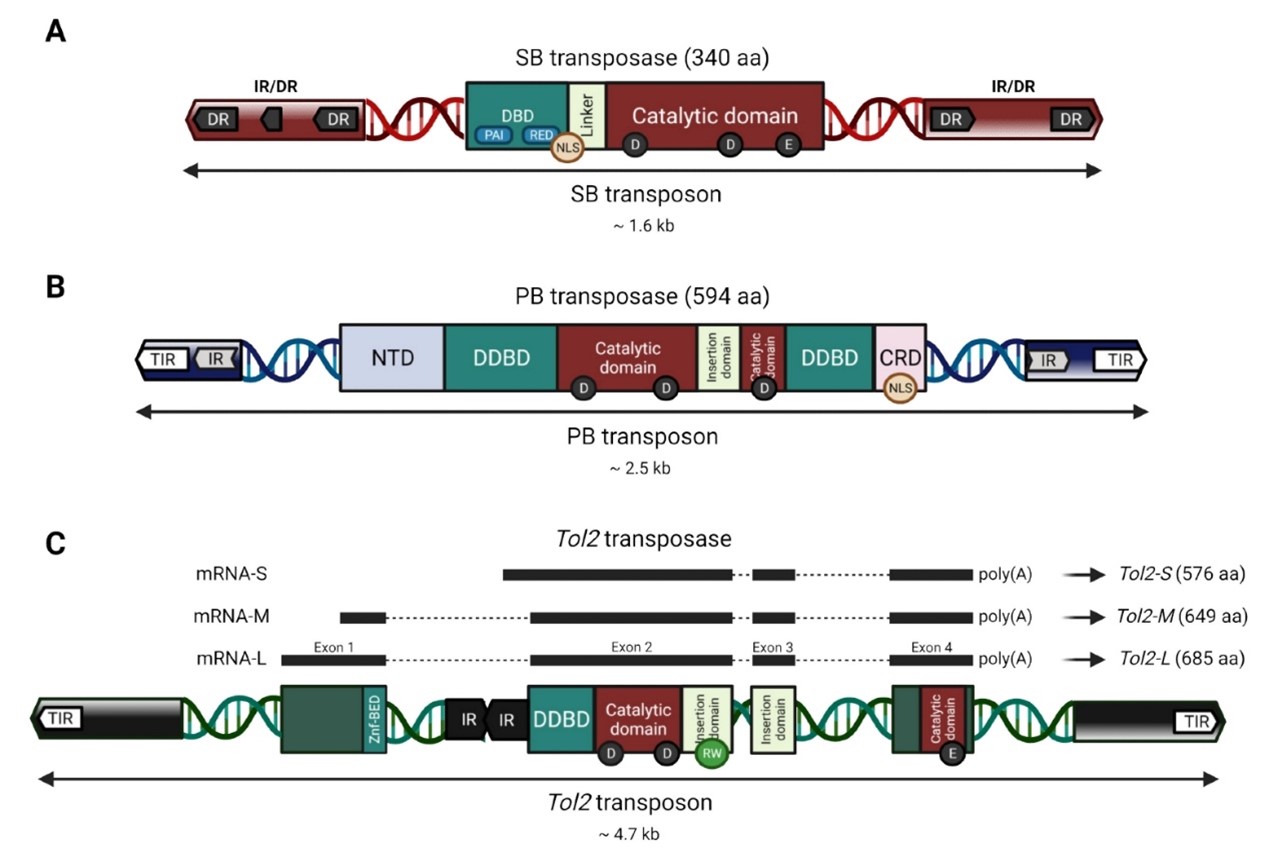
Fig 6. SB, PB, and Tol2 transposons
The SB transposon consists of two parts: the transposase gene and a two-terminal inverted repeat sequence that can be recognized by the transposase gene. The structure is shown in Fig 6A. The open reading frame of the transposase gene encodes a 340 amino acid protein that can bind to both ends of the inverted repeat sequence to facilitate transposition. The two-end reverse repeat sequence is about 230 bp long and consists of three parts: the outer 32 bp inverted repeat (IR), the inner homologous repeat (DR) similar to the IR, and a fragment 165 to 166 bp apart between the two. The DR is the binding region of the transposase and is a tightly conserved region of the transposon. In general, SB transposons integrate specifically into TA dinucleotide sites in the genome.
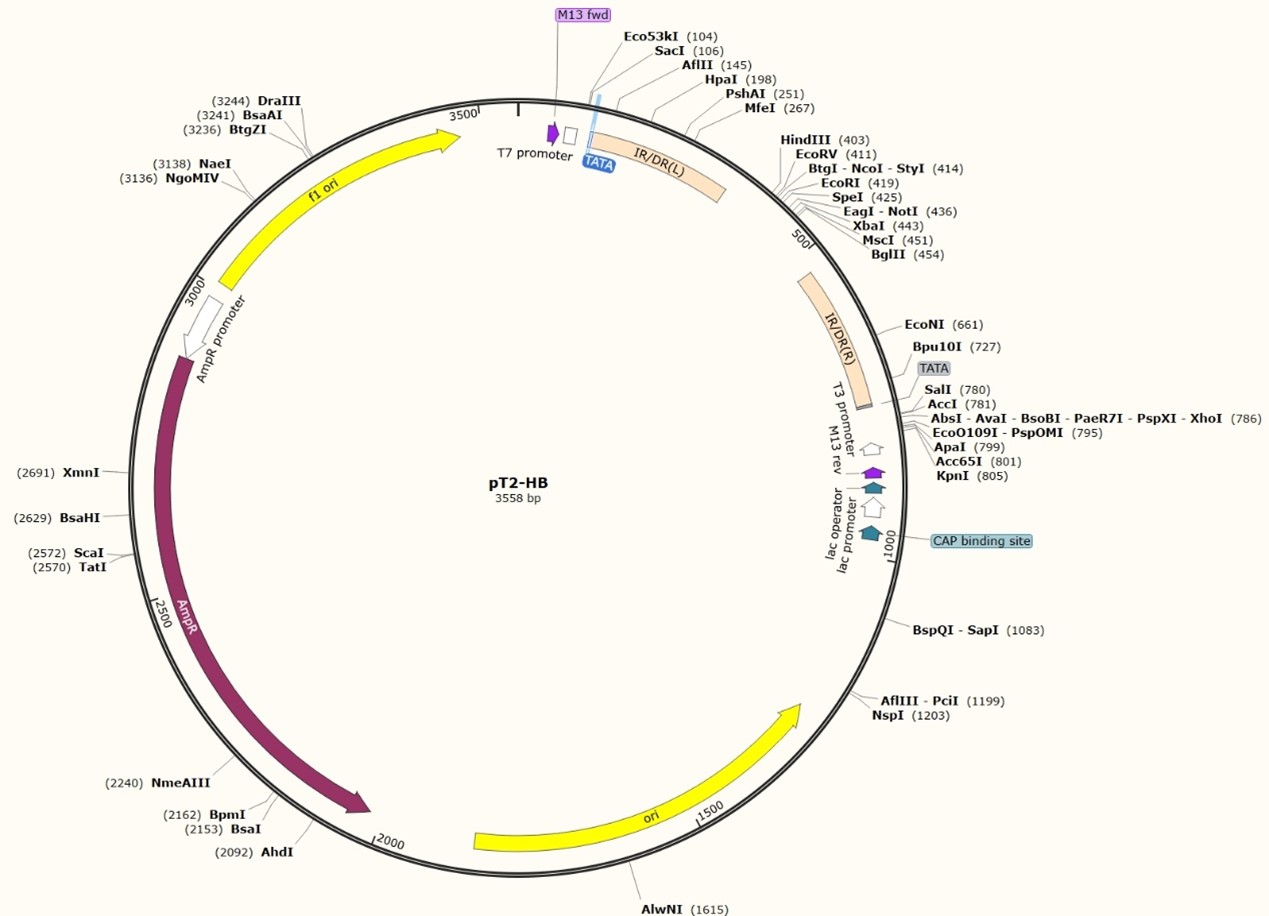
Fig 7. A typical SB transposon plasmid map
In the presence of SB transposase, IR/DR(L) and IR/DR(R) and the DNA fragments between them are integrated into TA sequence-containing sites in the genome.
Tol2 is an autonomous transposon element discovered in the genome of medaka fish. The structure of the Tol2 transposon is shown in Fig 6C. It encodes a transposase that catalyzes the transposition reaction of the 200 bp sequence at the 5' end and the 150 bp sequence at the 3' end in the Tol2 transposon structure.
The Tol2 transposon is undirected, which means it is inserted randomly in the genome and has no sequence specificity. The Tol2 system consists of two vectors: the plasmid responsible for encoding the transposase and the transposon plasmid. The transposon plasmid contains two inverted terminal repeats (ITRs) and the transposon region between them. The transposase recognizes the two ITRs of the transposon and inserts the transposable region and the two ITR elements into the host genome. Tol2 integrates genes by a "cut-and-paste" mechanism, generating an 8 bp repeat sequence at each insertion site.
| Sleeping Beauty (SB) | piggyBac (PB) | Tol2 | |
| Origin and Sources | Salmonid fish | Cabbage looper moth | Medaka fish |
| Classification | Tc1/mariner superfamily | PB superfamily | hAT superfamily |
| Length of Transposable element | -1.6kb | -2.5kb | -4.7kb |
| Terminal region | ~230bp of IRs/DRs | 35-63bp with external TIRs and internal subterminal IRs | 150-200bp containing TIRs and subterminal regions |
| Transposase | 340aa | 594aa | 649aa (most active isoform) |
| Footprint | CAG | None | Variable |
| Target site preference | TA | TTAA | Weak consensus sequence TNA(C/G)TTATAA(G/C)TNA |
| Target site repeat | TA | TTAA | 8bp |
| Activity in species | Various vertebrates | Vertebrates, insects, plants, yeast | Various vertebrates |
| Efficiency in human cells | Comparable to retroviral vectors | Comparable to retroviral vectors | Lower than PB and SB |
| Loading capacity | >100kb | >100kb | >100kb |
| Overproduction inhibition | Yes | To some extent | Lower than PB and SB |
| Integration profile | Nearly random | Favors TSSs, CpG islands, and DNasel hypersensitive sites | Favors TSSs, CpG islands, and DNasel hypersensitive sites |
| Most common vector backbone | pT2 | pXL-Bacll | pTol2, miniTo/2 |
| Most active transposase | hySB100X | hyPB | hTol2-M |
| Transposon delivery vectors | Plasmid DNA, pFAR, MC, non-integrating viral vectors, nanoparticles | Plasmid DNA, dbDNA, non-integrating viral vectors, nanoparticles | Plasmid DNA |
| Transposase delivery vector | Plasmid DNA, mRNA, SNIM RNA, recombinant protein (hsSB), non-integrating viral vector, nanoparticles | Plasmid DNA, mRNA, non-integrating viral vector, nanoparticles | Plasmid DNA, mRNA, recombinant protein (His-Tol2) |
| Clinical trials | Yes | Yes | No |
References
Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013 Jan;14(1):49-61. doi: 10.1038/nrg3374. PMID: 23247435.
Reznikoff WS. Tn5 as a model for understanding DNA transposition. Mol Microbiol. 2003 Mar;47(5):1199-206. doi: 10.1046/j.1365-2958.2003.03382.x. PMID: 12603728.
Sandoval-Villegas N, Nurieva W, Amberger M, Ivics Z. Contemporary Transposon Tools: A Review and Guide through Mechanisms and Applications of Sleeping Beauty, piggyBac and Tol2 for Genome Engineering. Int J Mol Sci. 2021 May 11;22(10):5084. doi: 10.3390/ijms22105084. PMID: 34064900; PMCID: PMC8151067.
Souce: NovoPro 2024-08-24
