Typically, TEV protease is thought to require Glycine (Gly) or Serine (Ser) at the P1' position for effective cleavage, but this could lead to non-native residues being present on the N-terminus after tag removal, potentially affecting protein activity.
To explore this, the authors constructed 20 variants of a fusion protein substrate, each with a different amino acid in the P1' position, and measured how efficiently TEV protease processed each variant in vivo and in vitro. Kinetic parameters such as Km and kcat were also evaluated for some peptide substrates[1].
Key findings include:
TEV protease is more flexible than previously thought, tolerating a variety of side chains at the P1' position with little impact on cleavage efficiency.
Some residues (like Proline) significantly reduce the efficiency, but many others (such as Met, Ala, and Cys) are well-tolerated (Fig 1.).
This flexibility implies that TEV protease can be used to produce recombinant proteins with nearly any N-terminal residue except Proline, expanding its utility for high-throughput purification and structural studies.
The results suggest that TEV protease can be reliably used to cleave fusion proteins without introducing unwanted N-terminal residues that could affect the protein's function.
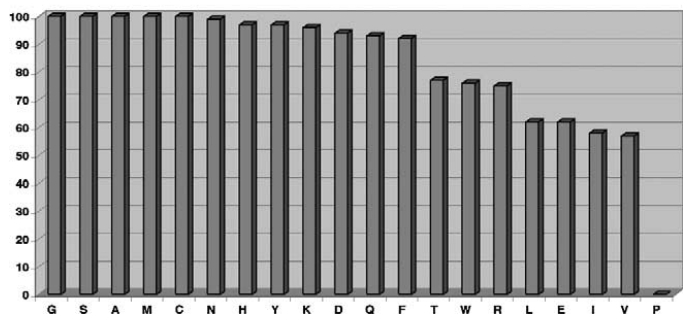
Fig 1. TEV protease cleavage efficiency of MBP-NusG with 20 different amino acids located at position P1′.
In another paper[2] the authors also investigate how various amino acids at the P1' position (the residue immediately following the cleavage site) affect the efficiency of cleavage by TEV protease.
Cleavage Efficiency: The study finds that TEV protease can accommodate most amino acids at the P1' position with little to no significant impact on processing efficiency, except for proline, which drastically reduces the efficiency of cleavage.
Specific Amino Acids: Although most residues are tolerated, certain amino acids, such as tryptophan (Trp), threonine (Thr), leucine (Leu), glutamate (Glu), arginine (Arg), aspartate (Asp), valine (Val), and isoleucine (Ile), reduce the cleavage efficiency to about 60% or less compared to others. For these residues, TEV protease cleaves less efficiently, and adjustments to cleavage conditions might be needed (Fig 2.).
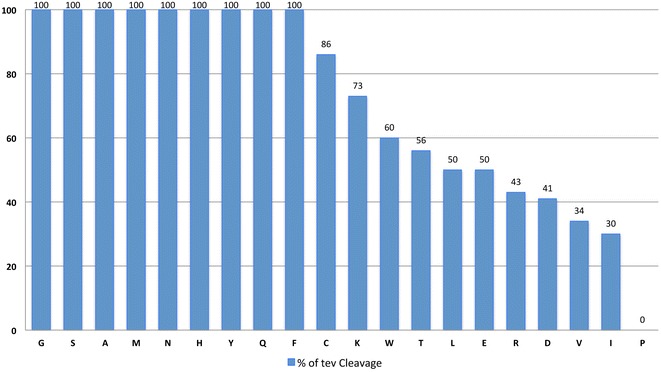
Fig 2. TEV protease cleavage efficiency of Kin17 with 20 different amino acids located at position P1′.
Confirmation: The study confirms that the previous understanding about proline being problematic for TEV cleavage holds true. However, for other amino acids, TEV protease generally retains some level of activity.
This implies that TEV protease is quite flexible with respect to the P1' position, making it versatile for protein processing applications.
References
- 1. Kapust RB, Tözsér J, Copeland TD, Waugh DS. The P1' specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002 Jun 28;294(5):949-55. doi: 10.1016/S0006-291X(02)00574-0. PMID: 12074568.
- 2. Sequeira AF, Turchetto J, Saez NJ, Peysson F, Ramond L, Duhoo Y, Blémont M, Fernandes VO, Gama LT, Ferreira LM, Guerreiro CI, Gilles N, Darbon H, Fontes CM, Vincentelli R. Gene design, fusion technology and TEV cleavage conditions influence the purification of oxidized disulphide-rich venom peptides in Escherichia coli. Microb Cell Fact. 2017 Jan 17;16(1):4. doi: 10.1186/s12934-016-0618-0. PMID: 28093085; PMCID: PMC5240416.
Souce: NovoPro 2024-10-12
