RNA splicing
RNA splicing is a necessary step in the post-transcriptional processing of RNA in eukaryotes and a key mechanism underlying the diversity and complexity of the transcriptome. Eukaryotic gene sequences contain introns and exons, which are interspersed. Introns are removed by the spliceosome after the gene is transcribed into pre-mRNA, leaving exons as the fragments retained in mature mRNA (which are subsequently translated into proteins).
To elucidate the molecular mechanisms of nuclear intron splicing, it is critical to understand the characteristics of splice sites. By comparing mRNA sequences with their corresponding genomic DNA sequences, it was found that the two ends of introns lack strong homology or complementarity. However, splice site sequences, though very short, exhibit highly conserved consensus sequences (as shown in Figure 1). At each consensus position, the height of each letter represents the percentage occurrence of a specific base. Thus, at the immediate boundaries of introns, highly conserved sequences can be identified. The canonical intron sequence is: GU…AG.
Since the recognition of splice sites at intron boundaries relies on the dinucleotides GU (start) and AG (end), this feature is termed the GU-AG rule (corresponding to GT-AG on the DNA coding strand).
Due to the distinct sequences of the two splice sites, they define the orientation of the intron. Moving left to right along the intron, the left boundary is called the 5' splice site (5'SS), also known as the *left splice site* or *donor site* (SD). The right boundary is the 3' splice site (3'SS), also termed the *right splice site* or *acceptor site* (SA). Mutations in splice sites block splicing in vivo and in vitro, further confirming the role of these consensus sequences in splicing.
In addition to the major introns governed by the GU-AG rule, organisms possess rare intron exceptions with distinct consensus sequences at exon-intron boundaries (Figure 1, lower panel). These were originally classified as minor introns under the AU-AC rule, as they conserve AU-AC dinucleotides at their boundaries (Figure 1, middle panel).
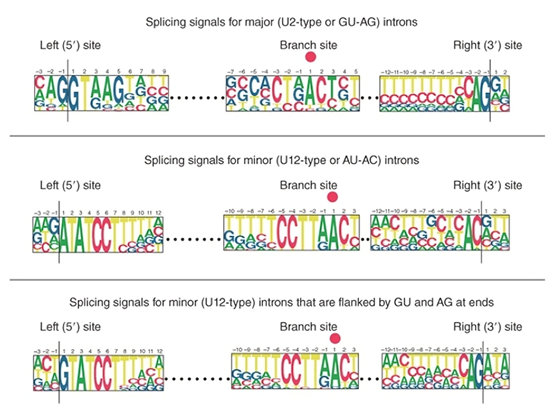
Figure 1. Structural and sequence features of major (U2) and minor (U12) introns.
Basic Process of Intron Splicing (Figure 2):
1. Cleavage of the phosphodiester bond between the exon and the 5' GU of the intron.
- snRNP U1 binds the 5' splice site via complementary sequences.
2. snRNP U2 binds the branch site, recruiting U1. The free 5' G forms a lariat structure via a phosphodiester bond with the branch site adenine (A).
3. Cleavage of the phosphodiester bond between the second exon and the 3' AG of the intron.
4. The intron is excised, and the two exons are ligated.
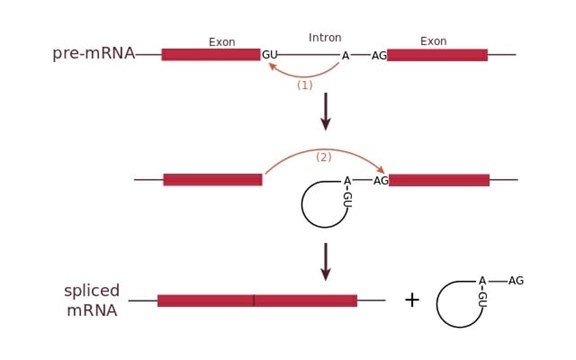
Figure 2. The splicing process of introns.
Exon Trapping
Exon trapping is a method to identify and capture transcribed sequences (exons) by leveraging the RNA splicing machinery to integrate and trap target exons into known genomic fragments, effectively identifying potential genes.
Procedure:
1. Cloning and Vector Construction:
- The genomic DNA fragment of interest is cloned into a modified vector containing a splice donor site (SD) and a multiple cloning site (MCS). The target DNA is inserted into the MCS.
2. Transfection and Expression:
- The vector is transfected into mammalian cells for transcription and splicing. If the inserted DNA contains a functional splice acceptor site (SA), splicing occurs, removing introns.
3. Exon Capture:
- Spliced RNA is reverse-transcribed into cDNA, which carries the trapped exon. The cDNA is amplified via PCR or other methods to analyze exon-donor site junctions.
4. Screening and Analysis:
- DNA sequencing or hybridization verifies exon trapping events. Valid events show correct exon-donor site junctions, indicating the exon is part of a gene.
This technique excels at identifying coding regions (exons) from long genomic DNA stretches, particularly in unannotated regions.
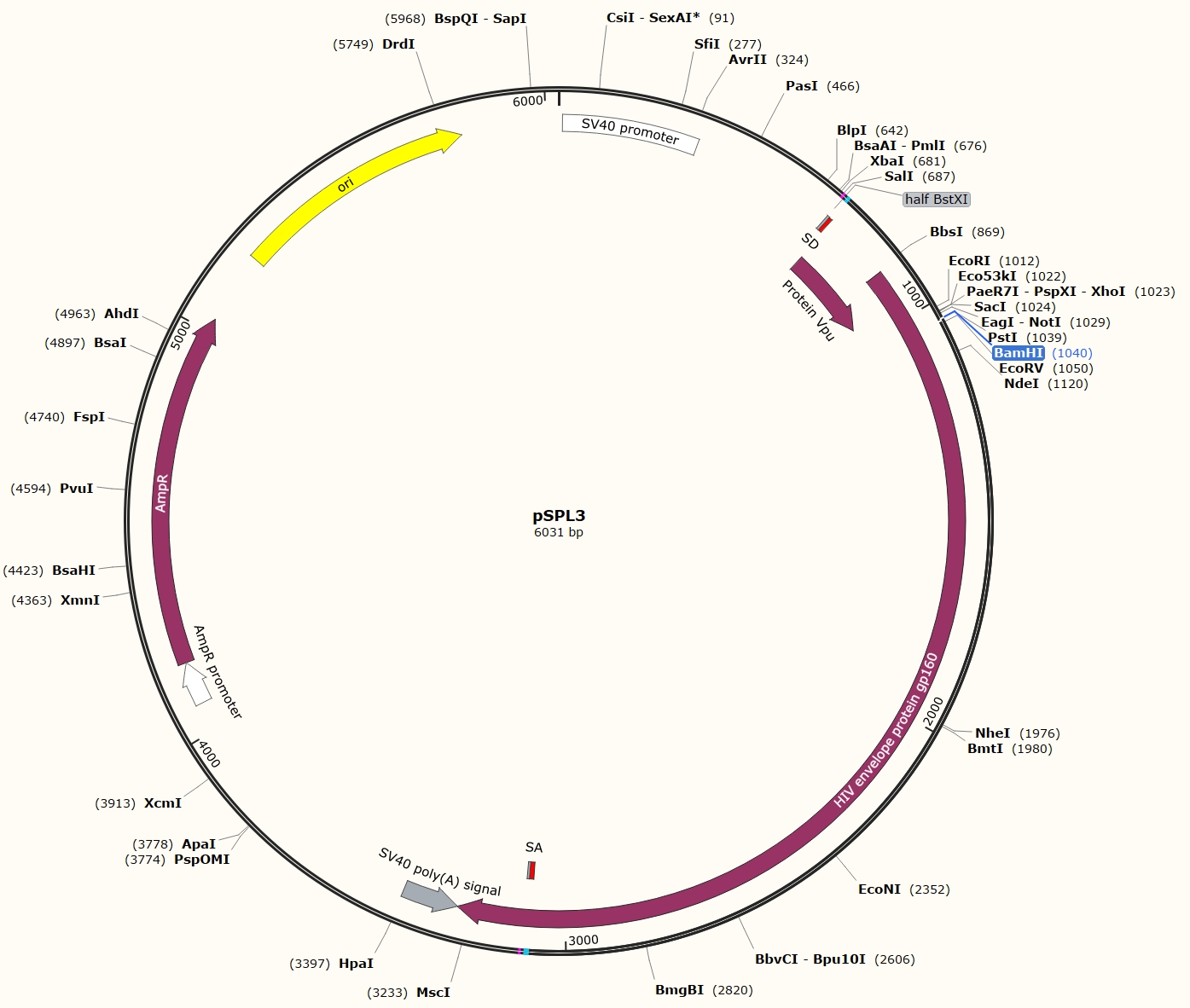
Fig 3. Map of pSPL3
Case Study from a Paper 1:
The authors constructed the plasmid pSPL3 (schematic below), which includes:
- SV promoter and polyA signal for transcription and polyadenylation in mammalian cells.
- SD and SA splice sites.
- Half BstXI sites: *ccagca* and *acctgg*. BstXI recognizes CCANNNNNNTGG. The HIV gp160 region contains multiple BstXI sites.
- BamHI site: Located between SD and SA; compatible with BglII (both produce *gatc* sticky ends).
Experimental Steps:
1. Digestion and Ligation:
- Genomic DNA is digested with BamHI and BglII.
- pSPL3 is digested with BamHI, dephosphorylated (to prevent self-ligation), and ligated to genomic fragments.
- Ligated products are transformed into *E. coli*, and plasmids are extracted for transfection into COS7 cells.
2. RNA Processing and PCR:
- mRNA is extracted 48–72 hours post-transfection, reverse-transcribed into cDNA, and amplified via PCR.
- To eliminate vector-only or false-positive products, PCR products are digested with BstXI and reamplified.
3. Cloning and Sequencing:
- PCR products are digested with SalI and BglII, then cloned into SalI/BamHI-digested pBluescriptIIKS+. Clones are sequenced and analyzed.
When genomic fragments contain intact exons with flanking intronic sequences, the exon is retained in mature polyA+ mRNA. RNA-based PCR detects the exon, enabling its capture.
References
Church DM, Stotler CJ, Rutter JL, Murrell JR, Trofatter JA, Buckler AJ. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. *Nat Genet*. 1994 Jan;6(1):98-105. doi: 10.1038/ng0194-98. PMID: 8136842.
Souce: NovoPro 2024-09-21
