Chemical genetics refers to the modification of biological macromolecules to interact with previously unrecognized small molecules, thereby achieving controlled and reversible (initiating or interrupting specific reactions with the addition or removal of compounds) regulation of the activity of these macromolecules, and specifically influencing physiological activities.
Chemical genetics techniques have been widely applied in research areas such as signal transduction, drug development, and functional genomics. The Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) technology is currently the most widely used chemical genetics technique. It was invented by Armbruster and Roth et al. in 2007. They altered the structure of the G protein-coupled receptor - the muscarinic acetylcholine receptor (mAChR) - so that it could only be activated or inhibited by a specific compound, Clozapine-N-oxide (CNO). These modified receptors selectively act on different GPCR cascade reactions. The most widely used ones are Gq-DREADD and Gi-DREADD, which are widely used to enhance or inhibit neuronal activity in a cell-specific and non-invasive manner.
Under normal physiological conditions, the human muscarinic acetylcholine receptor subtype M3 (hM3) binds to the endogenous neurotransmitter acetylcholine (Ach), and then couples with G protein-coupled receptors of the Gq class to participate in the Gq class signaling pathway. The human muscarinic acetylcholine receptor subtype M4 (hM4) binds to acetylcholine (Ach), and then functions by coupling with G protein-coupled receptors of the Gi class to participate in the Gi class signaling pathway. However, when two conserved sites on hM3 and hM4 were mutated (Y149C and A239G, and Y113C and A203G, respectively), both no longer bound to acetylcholine but instead efficiently bound to the exogenous CNO. We call the two mutated receptors hM3Dq and hM4Di, respectively. hM3Dq serves to excite neurons in response to CNO stimulation, while hM4Di leads to the inhibition of neuronal cellular action potential firing.
Some additional basics in cell biology: The physiological functions performed by GPCRs are largely determined by the intracellular effector proteins (such as G proteins) that they activate. G proteins are heterotrimeric proteins composed of Gα, β, and γ subunits. Ligand-bound GPCR induces conformational changes in the Gα subunit, resulting in guanine nucleotide exchange (GDP release and GTP binding) and dissociation of the Gα subunit from the Gβγ complex. The Gα subunit is a major determinant of GPCR signaling. Based on downstream signaling and sequence similarity, the 16 Gα subunits encoded in the human genome are divided into four subclasses: Gs, Gi, Gq, and G12. Each subclass elicits unique responses, such as an increase in cAMP (Gs), a decrease in cAMP (Gi), an increase in Ca2+ and IP3 (Gq), and activation of Rho GTPase (G12). Both hM3 and hM4 mentioned above are GPCRs that participate in different signaling pathways.
Adeno-associated virus (AAV) is widely used in biology, especially neurobiology, as a safe, long-lasting, efficient, and highly specific tool for gene manipulation. AAV is one of the most important gene delivery vectors for gene therapy. To specifically regulate gene expression in nerve cells, methods such as in situ injection, specific serotypes, specific promoters, and the Cre-loxP system can be used, either individually or in combination. For example, the simultaneous use of in situ injection + specific serotypes + specific promoters provides triple assurance for the specific regulation of genes in vivo.
AAV serotypes such as AAV1, AAV2, AAV4, AAV5, AAV8, and AAV9 can target the central nervous system (CNS); the hSyn promoter can initiate transcription specifically in mature neurons, CaMKIIα is specifically expressed in excitatory neurons (i.e., glutamatergic neurons), the GFAP promoter is specifically expressed in astrocytes, and CD68 is specifically expressed in microglia. For the Cre-loxP system, we can adopt a conditional gene activation expression strategy. At the beginning, the target gene is by default not expressed, but it will be activated and expressed in the presence of Cre recombinase co-expression, and the coding direction of the target gene will be reversed and changed. In the presence of Cre, the expression of the target gene is regulated by a user-defined promoter. Let's focus on the Cre-loxP system.
Common Ways to Activate Cre-dependent Genes
There are two common ways to activate Cre-dependent genes: lox-STOP-lox and DIO/FLEX switching (as shown in Figures 1 and 2 below).
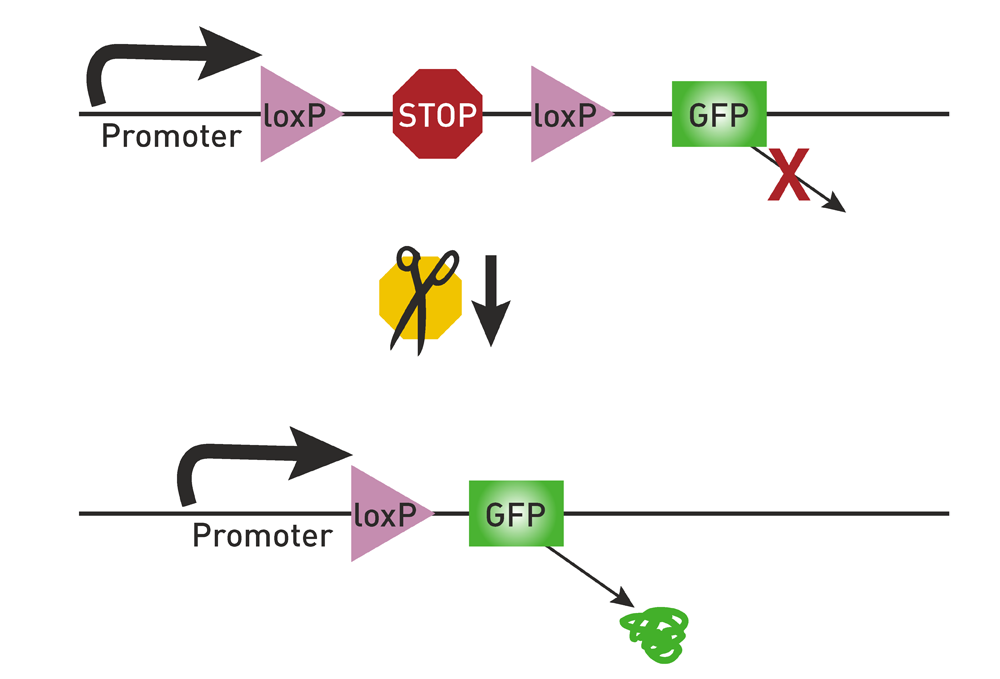
Fig 1. lox-STOP-lox
Before the target gene (such as the green fluorescent protein GFP in the figure below), a gene sequence containing two isotropic Loxp is constructed. Between the two Loxp is a STOP sequence that prevents the gene from being transcribed. Due to the presence of the STOP sequence, the green fluorescent protein cannot be transcribed and translated. When Cre appears, Cre will excise the STOP sequence between the homologous Loxp. After that, the green fluorescent protein will be successfully expressed. This process is reversible. The lox-STOP-lox strategy has the disadvantage of leaky expression.
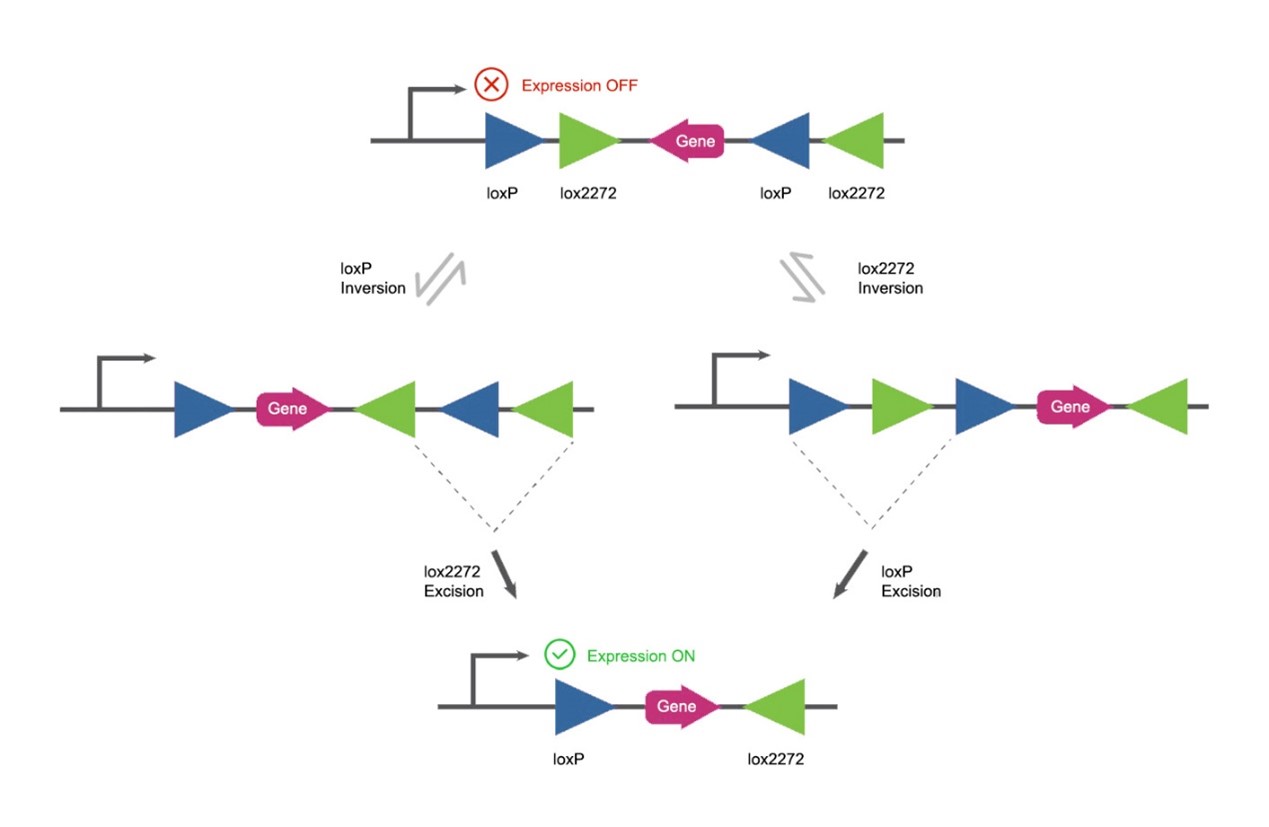
Fig 2. DIO/FLEX
The sequence between the two LoxP is the target gene, and the sequence of the target gene is reversed and cannot express a meaningful protein. When Cre appears, the successive recombination action of Cre will eventually change the reversed target gene into a positive one, after which the target gene can be normally expressed.
The recombination process of Cre is a two-step process:
The first step is inversion. Under the action of Cre, the sequence between two LoxPs with opposite directions will be inverted. The inversion may occur between two LoxPs or between two Lox2722s. Whichever occurs, it will not affect the final result. This step of the recombination reaction is reversible.
After the inversion occurs, the next step is Cre-triggered recombination deletion. The inversion triggered by LoxP will change the two otherwise reversed Lox2722 to isotropic and vice versa. The two LoxP or Lox2722 sites in the same direction will trigger the shearing function of Cre. Cre will shear off one Lox2722 and one LoxP. There will be no more recombination of Cre between the last remaining LoxP and Lox2722, and it will not be reversed. It is a stable state.
Cre recombinase can be introduced by either AAV or Cre tool mice; the Cre tool mice mainly fall into three categories: systemic Cre, tissue-specific Cre, and ligand- or drug-induced Cre.
We will integrate the above content using a paper [1] as a case study.
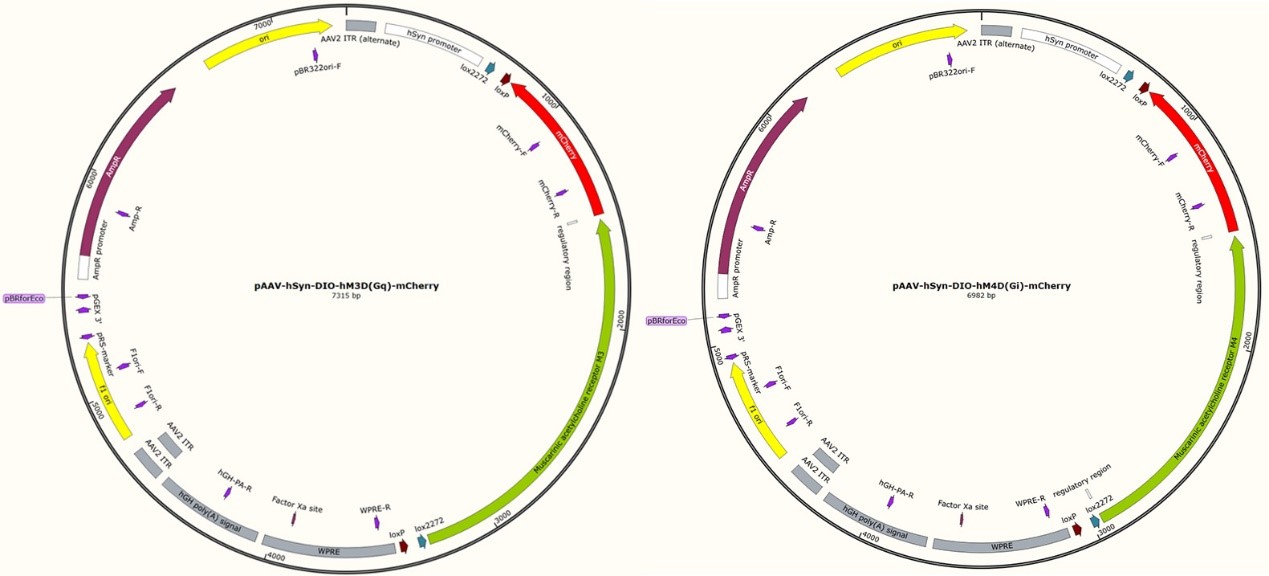
Fig 3. Mapping of the two plasmids used in the paper
Basic Experimental Procedures:
1. Construct the two plasmids described above to express hM3Dq and hM4Di. The authors used a DIO strategy: only in the presence of Cre recombinase, the mature neuron-specific hSyn promoter can initiate the transcriptional expression of hM3Dq and hM4Di.
2. Package AAV viruses together with the RepCap plasmid of the AAV8 serotype.
3. Inject the AAV virus into the brains of mice (AgRP-Ires-cre) in which AgRP neurons specifically express Cre recombinase by stereotaxic in situ injection.
4. Inject CNO to achieve control of neuronal activity.
5. Conduct electrophysiological assays: use electrodes to record the voltage changes inside and outside the neuronal cell membrane to verify the effectiveness of the DREADDs receptor.
6. Perform phenotypic assays: experimentally confirm the effects of activating or inhibiting neuronal activity on experimental animals.
Reference
[1] Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL, Lowell BB. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011 Apr;121(4):1424-8. doi: 10.1172/JCI46229. PMID: 21364278; PMCID: PMC3069789.
Souce: NovoPro 2024-09-12
